Date Approved: August 21, 2024
Date of Last Update: August 25, 2025
Version: 3.1
List of Abbreviations
AnVIL: Genomic Data Science Analysis, Visualization, and Informatics Lab-space
CC: PRIMED Coordinating Center
CDSA: Consortium Data Sharing Agreement
DAC: Data Access Committee
DAR: Data Access Request
dbGaP: The Database of Genotypes and Phenotypes
DSP: Data Sharing Policy
DUL: Data Use Limitations
EL: PRIMED Eligibility List
GDS: Genomic Data Sharing
GSR: Genomic Summary Results
GWAS: Genome-Wide Association Studies
IRB: Institutional Review Board
NCI: National Cancer Institute
NHGRI: National Human Genome Research Institute
NIH: National Institutes of Health
PGS: Polygenic Score
PRIMED: Polygenic Risk Methods Development
PRS: Polygenic Risk Score
SAG (or PRIMED-SAG): PRIMED Consortium Data Sharing Agreement Group, see also Definitions
Definitions
- Data sharing circle: a group of investigators with permission to access the same data such that the data can be shared among them collectively.
- Genomic summary results (GSR): previously referred to as ‘aggregate genomic data’ or ‘genomic summary statistics,’ are generated from primary analyses of genomic research. They convey information relevant to genomic associations with traits or diseases across datasets rather than associations specific to any one individual research participant. GSR include systematically computed statistics such as, but not limited to: 1) frequency information (e.g., genotype counts and frequencies, or allele counts and frequencies); and 2) association information (e.g., effect size estimates and standard errors, and p-values). Source: NIH Guide Notice NOT-OD-19-023.
- PRIMED Consortium Data Sharing Agreement Group (PRIMED-SAG or SAG) refers to the data sharing circle established by CDSA signees.
PRS* Terms and Definitions
| PRS Term and Definition | Data Classification |
|---|---|
|
PRS method / algorithm: analytical/statistical approach to analyze individual- or summary-level data and generate a new PRS model
|
Not data |
|
PRS model
|
Genomic Summary Results (GSR)** Summary-level data (not GSR); reported in publications |
|
Individual PRS: the output numeric value calculated from a PRS model for an individual. This may be a raw score or may be presented as an adjusted score in the context of a population distribution – i.e. as a percentile or number of standard deviations from the mean.
|
Individual-level data |
|
PRS performance metrics: numerical metrics describing performance of the PRS model in some population(s)
|
Summary-level data (not GSR); reported in publications |
|
*Terms “PRS,” “PGS,” and “GRS” (genetic or genomic risk score) can be used interchangeably for these definitions in this policy context (see also Wand et al. 2021 for distinctions); may also be referred to as “PGI” (polygenic index) in some disciplines (Burt et al. 2024) **GSR derived from one or more studies with a sensitive designation can only be shared via controlled access. See Data Sharing outside of PRIMED for further information. †See the PRIMED Recommendations for use of population descriptors (linked under the Resources section of the POPSEI WG page) |
Scope
To facilitate access, sharing, and broad usage of datasets with genomic and health measures from diverse ancestry populations within the PRIMED (Polygenic Risk Methods Development) Consortium, the PRIMED Consortium, NHGRI, and NCI have established the Data Sharing Policy (DSP) herein presented. The scope of this policy extends to the expectations and timelines for bringing in existing and/or newly generated data from dbGaP, PRIMED Study Sites and other data contributors. The processes and procedures outlined in the DSP are designed to promote scientific efficiency, synergy, and collaboration by facilitating Consortium-wide cross-study analyses, while protecting study participants’ privacy and respecting consent.
The DSP addresses intra-Consortium sharing of contributed individual-level data and summary-level data, as well as derived data products generated in PRIMED. Two initial mechanisms for intra-Consortium sharing are described: Coordinated dbGaP Applications and a Consortium Data Sharing Agreement (CDSA). The DSP also addresses appropriate sharing or release of derived data products generated in PRIMED to the scientific community.
It is incumbent upon PRIMED investigators to be aware of and follow additional policies that apply to their activities, whether within or beyond the scope of this PRIMED DSP. Such additional policies include the National Institutes of Health Genomic Data Sharing Policy (GDS), Consortium Guidelines for AnVIL Data Access, the Genomic Data User Code of Conduct, and any other local or institution-specific policies.
Back to topPolicy
Eligibility for Consortium Data Access and Sharing
In order to enter into PRIMED Consortium-wide data sharing circles, an investigator’s name must be included on the PRIMED Eligibility List (EL). Specifically, submitted PRIMED Coordinated dbGaP applications will be disapproved by the NIH Data Access Committees (DACs) if the applicant’s name does not appear on the EL. Similarly, signed CDSAs will not be accepted by the Coordinating Center if the investigator representative (signatory) is not on the EL. PRIMED PIs are listed on the EL by default and should contact the CC to add PRIMED co-Investigators. PRIMED Affiliate Members have the opportunity to enter Consortium data sharing circles; see the Affiliate Membership Policy for details.
The PRIMED Eligibility List includes:
- Contact PIs and multiple PIs of all Study Sites and the Coordinating Center
- Co-Investigators of Study Sites and the Coordinating Center, upon request from a contact or multiple PI
- Affiliate Group PI, upon indication and approval to enter PRIMED Consortium-wide data sharing circles during the Affiliate Membership application process and Steering Committee approval
- Co-Investigators of Affiliate Groups, upon request from an Affiliate Group PI
- PIs, multiple PIs, co-investigators of multi-institutional Affiliate Groups (e.g. CDSA DATA AFFILIATE COMPONENT groups), upon request from an Affiliate Group PI (e.g. Primary not COMPONENT PI)
Note that EL members do not gain access to Consortium datasets until they are approved via the relevant data access mechanism (e.g., DACs have approved relevant Data Access Requests on a PRIMED coordinated dbGaP application, or a signed and executed CDSA has been submitted to the Coordinating Center).
PRIMED data is stored, shared, and accessed via the AnVIL platform. All Consortium members must agree to abide by the Consortium Member Responsibilities in the Consortium Guidelines for AnVIL Data Access, including establishing Two Factor Authentication on their Google Account, in order to be granted access.
Additional eligibility requirements specific to coordinated dbGaP applications are provided under PRIMED Coordinated dbGaP Applications below.
Mechanisms of Consortium Data Access and Sharing
Due to the volume and heterogeneity of data and data sources used in PRIMED, multiple sharing mechanisms are needed. Two primary mechanisms are described below. Additional coordinated access mechanisms may be added over the life of the Consortium to enable access and sharing of additional controlled-access datasets with requirements that are unique or otherwise unsatisfied by the two primary mechanisms, such as for the UK Biobank (UKBB), eMERGE Network, All of Us Research Program (AoU), and the VA Million Veteran Program (MVP). Note that open/unrestricted access data can be shared within the PRIMED Consortium without a coordinated access mechanism (see Data Management and Access for more information).
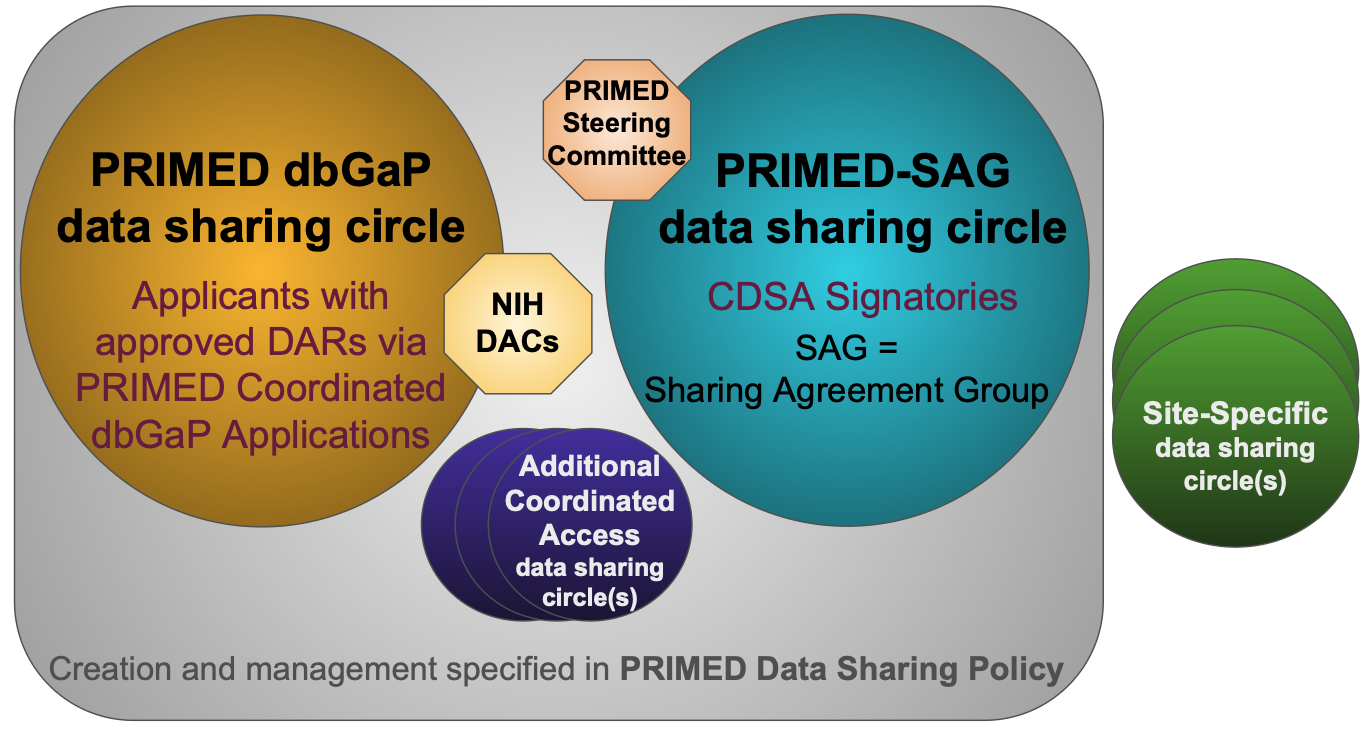
PRIMED Coordinated dbGaP Applications
PRIMED is using coordinated dbGaP applications to create a sharing circle for controlled-access data released by or otherwise accessible via dbGaP (gold circle in Figure 1). Applications with the same title, Research Use Statement, collaborator list, and other key elements can be used to allow investigators across institutions to share data (i.e. investigators not otherwise eligible to be covered by a single application). Data for a given study-consent group can only be shared among applicants with approved Data Access Requests (DARs) to that given study-consent group. This process was modeled off of a precedent from the NHLBI Trans-Omics for Precision Medicine (TOPMed) program. PRIMED investigators should follow the Instructions for PRIMED Coordinated dbGaP Applications to apply. Approval is expected if the PRIMED-specific process is followed completely.
Prior to submitting an application, applicants should review information about the datasets they will be requesting to see if the following are needed. Obtaining this documentation may take time:
- If requesting datasets that have a consent group that contains an Institutional Review Board (IRB) modifier, applicants must obtain local (i.e. from their institution) IRB approval for their proposed PRIMED analyses.
- If requesting datasets that have a consent group that contains a Collaboration Required (-COL) modifier, the applicant must provide a letter of collaboration with the primary study investigator(s). The letter of collaboration must be renewed every year. Refer to Letter of Collaboration templates.
To initiate a dbGaP application, the following prerequisites must be met:
- The applicant must be eligible to apply to access dbGaP data: dbGaP Authorized Access Portal > click on “Who can apply for access?”
- The applicant must apply to access dbGaP data: dbGaP Authorized Access Portal > click on “How does one apply?”
- The applicant’s dbGaP account must be in good standing.
Separate access requests must be filed per institution, even if collaborators are within the same funded Study Site within PRIMED. When a study investigator moves to a new institution, s/he must submit a new request from that institution. Data obtained via application from one institution may not be transferred to another institution. The PRIMED Eligibility List is appended to each PRIMED application to indicate external collaborators (i.e., collaborators at a different institution than the applicant) with whom data can be shared via the coordinated applications - see Data Management and Access below.
PRIMED Consortium Data Sharing Agreement (CDSA)
The PRIMED Consortium Data Sharing Agreement (CDSA) enables intra-Consortium sharing of studies and datasets provided by PRIMED Study Sites and/or Affiliate Member data contributors. The CDSA therefore creates an additional sharing circle, the PRIMED Consortium Data Sharing Agreement Group, or PRIMED-SAG (teal circle in Figure 1) and enables sharing of additional data (e.g., data not available via dbGaP). The PRIMED CDSA was modeled off precedents from the CHARGE and C4R consortia.
PRIMED Core Members do not require Steering Committee approval to join the PRIMED-SAG, either as MEMBERs or DATA AFFILIATES. PRIMED Affiliate Members do require Steering Committee approval to join the PRIMED-SAG, either as DATA AFFILIATES or NON-DATA AFFILIATES. COMPONENTS (associated centers or institutions who wish to use SAG Data) can join the PRIMED-SAG if they are represented by and have a relationship to their respective MEMBER, DATA AFFILIATE, or NON-DATA AFFILIATE. COMPONENTS do not require Steering Committee approval to join the PRIMED-SAG. However, each MEMBER, DATA AFFILIATE, NON-DATA AFFILIATE, and each of their COMPONENTS must submit a copy of the signed and executed Consortium Data Sharing Agreement (CDSA) to the PRIMED CC.
For those interested in signing the CDSA as a DATA AFFILIATE:
- If the PI who can sign the CDSA on behalf of the study/cohort/consortium is already a PRIMED Core Member, they will not need to apply for Affiliate Membership.
- If the PI who can sign the CDSA on behalf of the study/cohort/consortium is not already a PRIMED Core Member, they will need to apply for Affiliate Membership through the Data Affiliate application process (see Affiliate Membership Policy).
For those interested in signing the CDSA as a NON-DATA AFFILIATE:
- If the PI who can sign the CDSA on behalf of a research team/lab is already a PRIMED Core Member, they will not need to apply for Affiliate Membership.
- If the PI who can sign the CDSA on behalf of the group/research team is not already a PRIMED Core Member, they will need to apply for Affiliate Membership through the Expertise-only (Non-Data Affiliate) application process (see Affiliate Membership Policy).
See also definition of Affiliate Member Applicant in the PRIMED Affiliate Member Policy.
Data Management and Access
PRIMED Consortium data are to be uploaded, stored, and accessed on the NHGRI’s AnVIL cloud platform. Data in the PRIMED Consortium AnVIL workspaces will be accessible only by eligible PRIMED investigators who have been granted secure data access via the PRIMED Mechanisms of Access (i.e., Coordinated dbGaP Applications; signed and executed PRIMED Consortium Data Sharing Agreement). Data access permits data management and analysis in AnVIL workspaces. Downloads of data are not permitted.
Details on the management of PRIMED Consortium data – including data organization in AnVIL open/unrestricted and controlled-access workspaces, management of user access lists, expectations for data formatting, and the process for users to upload, validate, and access data – are described in detail in the PRIMED Data Management and Funding Plan and on the internal Consortium website, under “Getting Started with Data.” The plan also describes the distribution of cloud expenses, including data storage and computation, across the PRIMED Study Sites and Coordinating Center.
Access Removal, Withdrawal, and Termination
When Eligibility List members change institution and do not retain an appointment at their original (i.e. at the time of joining PRIMED) institution, the Coordinating Center has the role to remove reader/uploader access for a CDSA’s investigator REPRESENTATIVE (Eligibility List member) and their team after a three month grace period.
A dbGaP applicant is able to close out their PRIMED Coordinated dbGaP project application in order to leave the dbGaP data sharing circle. See the dbGaP procedure for Project Closeout.
For details pertaining to the terms and conditions of withdrawal, removal, and termination events in the PRIMED-SAG, please refer to the following articles of the CDSA:
- Terms and Conditions 7. Withdrawing from the PRIMED-SAG
- Terms and Conditions 8. Removing a MEMBER, AFFILIATE, or COMPONENT
- Terms and Conditions 12. Terminating the PRIMED-SAG
Further details on SAG management related to CDSA version updates are below.
SAG management
Below, “VN” refers to the current major version of the CDSA, “VN.K” to differing minor versions within the current major version, and “V(N-1)” to the previous major version.
- The SAG is formed by signatories of the current major version of the CDSA (VN). Differences in minor version (e.g, CDSA VN.K) are allowed in the SAG.
- A major version becomes current immediately upon approval by the PRIMED Steering Committee.
- Data from new DATA AFFILIATE signatories can be uploaded and made available upon signing the current major version.
- When incrementing a major version update of the CDSA (i.e. from V(N-1) to VN), signatories of the previous version have a three month grace period to sign the new version in order to remain in the SAG.
- Data uploaded during previous version(s) of the CDSA, i.e., V(N-1), remain available to pre-existing and new signatories during the grace period.
- Data uploaded during the grace period under CDSA VN would become available to pre-existing and new signatories during the grace period.
- For all signatory categories, failure to update to the current major version by the end of the grace period would result in removal from the SAG and loss of access to the data available in the SAG.
- For DATA AFFILIATES, failure to update to the current major version by the end of the grace period would further result in the removal of their data contribution from the SAG. An extension may be granted by the PRIMED Executive Committee if documentation of signature progress can be provided.
- There are two routes through which a signatory can update to the current major version of the CDSA (VN):
- (1) Have previously signed CDSA V(N-1) and sign the V(N-1) to VN Amendment
- For example when V3.0 is the current version, sign V2.1 and the V3.0 Amendment
- (2) Sign CDSA VN (or sign CDSA VN.K)
- For example when V3.0 is the current version, sign V3.0 or V3.K
- (1) Have previously signed CDSA V(N-1) and sign the V(N-1) to VN Amendment
- Future major version updates to the CDSA (beyond V3.0) are not anticipated at this time, given the multiple rounds of institutional feedback and consultation to arrive at V3.0 (and ~1.5 years after initial drafting). Major updates may be considered in the future, at the discretion of the PRIMED Executive Committee.
Data Sharing Guidelines and Expectations
Data Sharing within PRIMED
PRIMED Study Sites will share data with each other using the Consortium data sharing mechanisms defined above, and leverage AnVIL shared workspaces managed by the CC (see also PRIMED Data Management and Funding Plan). Data sharing applies to source data and any data that are derived, harmonized, imputed, or re-processed from those data. Existing summary level data (e.g., GWAS summary statistics, PRS weights, allele frequencies) that can be shared, will be shared within the Consortium in AnVIL. All individual participant or individual-level genotype and phenotype data that can be shared, will be shared within the Consortium in AnVIL. Some datasets may not be shared due to data use limitations (DULs) or restrictions around redistributing the data in AnVIL.
Data upload and sharing within the Consortium should (1) be prompt and supportive of the productivity of the Consortium’s output and (2) in alignment with PRIMED RFA and NIH program requirements and directives.
Data Sharing outside of PRIMED
Data generated within PRIMED will be shared with the broader scientific community whenever possible and in alignment with NIH data sharing policies and study-specific data use limitations. As a Consortium developing PRS methods based largely on secondary use of extant data, data generated by the Consortium is expected to fall into the following general categories:
- Genomic Summary Results: e.g. Association analysis (i.e., GWAS) results, allele frequencies
- PRS models and performance metrics (see also PRS Terms and Definitions)
- Individual-level: harmonized phenotype and genotype data, which is based on pre-existing source (i.e. unharmonized) data
- Individual-level: newly generated/collected phenotype and genotype data may to a lesser extent be generated in PRIMED, e.g., through the supplemental genotyping program
Note the sharing of Genomic Summary Results (GSR) in PRIMED will follow the 2018 Update to NIH Management of Genomic Summary Results Access (NOT-OD-19-023). Notably, this policy update defined the “sensitive” study designation, wherein an institution can determine that GSR from the given study require controlled access sharing. After consulting with the NHGRI Data Sharing Governance Committee, in PRIMED we are considering any component of a PRS model that contains data otherwise considered GSR to be itself GSR and thus falling under the NIH GSR sharing policies. For example, the list of variants with weights/effect sizes used to calculate the PRS (sometimes referred to as a "scoring file") are GSR; see the PRS Terms and Definitions section.
Below are opportunities for sharing these data generated by the Consortium with the broader scientific community, along with caveats.
- Newly generated summary-level data (e.g., GWAS summary statistics, allele frequencies, PRS models, other analysis outputs) along with associated documentation will be released outside of the Consortium on a platform/repository and under the access model appropriate for the given datasets:
- Summary-level data requiring controlled access may be registered via dbGaP and shared on the AnVIL platform, with specific data use limitations inherited from the source individual-level data. Specifically, “any data use limitations attached to use of the individual-level data will apply” (see NOT-OD-19-023).
- In PRIMED, “sensitive” designation is obtained from dbGaP study registration or the CDSA questionnaire.
- Following precedent from dbGaP study registration, a multi-study GSR derived from one or more sensitive studies will also be considered sensitive and require controlled access sharing. For multi-study GSR with multiple contributing consent groups, the most restrictive consent designation will apply.
- PRS models where the PGS scoring file requires controlled access will be shared via an appropriate repository or combination of repositories. The Consortium is investigating whether such PRS models may be registered in the PGS Catalog to display the PRS model metadata, with the PGS scoring file stored and accessed separately in a controlled access repository such as AnVIL.
- Open/Unrestricted access GWAS statistics may be deposited to the GWAS Catalog.
- Open/Unrestricted access PRS models may be deposited to the PGS Catalog.
- Per guidance received from NIH program staff and the NHGRI Policy Advisor, via discussion with NIH Office of Science Policy: A secondary data user registering a study to share controlled access GSR based on source data obtained from dbGaP does not violate the “non-transferability” clause of the dbGaP Data User Code of Conduct given that the data derivative being shared is summary- rather than individual-level.
- Newly generated individual-level data (e.g., harmonized phenotypes, imputed genotypes) derived from source data accessed via dbGaP applications may be shared outside the Consortium through the existing study accessions for the source studies, with specific data use limitations inherited from the source individual-level data.
- This requires obtaining permission from and working closely with the study owners/data generators.
- Pre-existing data obtained from publicly available open or controlled access sources will not be re-released and/or redistributed by the PRIMED Consortium.
PRIMED investigators with questions on sharing Consortium-generated data products with the broader scientific community should consult the Data Sharing Working Group, Coordinating Center, and/or PRIMED NIH program staff. PRIMED should leverage the opportunity to document potential gaps or lack of clarity in sharing policies, including potential solutions.
Data Use Guidelines
Each Consortium member agrees to comply with all limitations and use restrictions accompanying the PRIMED data or data accessed via the PRIMED Mechanisms of Access described above. Consortium members and other data contributors will convey any such limitations to other members that access these data through PRIMED sharing mechanisms. Limitations should appropriately reflect the informed consent of Study participants from whom the data shared under this agreement were collected and derived. Each data contributor or data generator will provide any limitations (e.g., cardiovascular disease research only, sharing of summary results requires controlled access) on use of the data they are sharing with the Consortium, and, when in doubt about the appropriateness of sharing, will consult with an ethics board or IRB. As applicable, all data use must be consistent with PRIMED policies, dbGaP approved data use limitations, and comply with the responsibilities for investigators accessing and using genomic data specified in the NIH Genomic Data Sharing Policy.
All PRIMED analyses and scientific activities within scope of the PRIMED Publications Policy require an approved proposal. Data obtained via the PRIMED Mechanisms of Access described above should therefore only be used under the auspices of an approved proposal.
dbGaP data
When study investigators apply for access to controlled access data (e.g., dbGaP), they will certify they and their team’s data uses will comply with the Data Use Limitations specified for each study and consent group. Data from an individual with a disease-specific consent will not be used in analyses outside of that restriction, unless specifically allowed by the Data Use Limitations. The applicant PI’s team includes anyone who has access to or uses data under the DAR approvals conferred in the applicant PI’s dbGaP project application. This includes those listed as Internal Collaborators, designated downloaders, as well as unlisted lab staff and trainees who are covered by NIH Policy as described in the dbGaP FAQ on Collaborators as well as the Instructions for PRIMED Coordinated dbGaP Applications section “Collaborators” tab.
PRIMED-SAG Data (via CDSA)
As documented in the PRIMED CDSA as well as the Instructions for PRIMED CDSA section Standard set of PI reminders regarding the use/analysis of SAG Data, the CDSA signees take on the responsibility to ensure they and their team members are using CDSA datasets in a way that complies with all data use permissions, modifiers, and limitations for both individual-level and summary-level data, as specified for a given dataset. The signee’s team includes anyone who has access to or uses data under their fully executed CDSA, including named data accessors.
For details pertaining to compliance with limitations accompanying the use of SAG Data, please refer to the following articles of the CDSA:
- Terms and Conditions 4. Complying with Consents
- Definitions 5. Representative
Additionally, any PRIMED Consortium member (or group of members) who wishes to make use of PRIMED-SAG Data would indicate the proposed studies/cohorts/consortia (DATA AFFILIATES who signed the PRIMED Consortium Data Sharing Agreement) when submitting a PRIMED paper proposal. The study/cohort/consortium contact (if they opt in) would get an email to review the paper proposal (approve/disapprove and leave comments regarding the use of their study’s data). This optional measure gives DATA AFFILIATES oversight over how their data is used within the PRIMED-SAG.
In order for CDSA COMPONENTS to obtain access/upload permissions to PRIMED-SAG Data, the Primary MEMBER or AFFILIATE to which the center is associated will first need to obtain such access/upload permissions to PRIMED-SAG Data. In other words, the Primary MEMBER, DATA AFFILIATE, or NON-DATA AFFILIATE will need to first join the PRIMED-SAG (via a fully executed PRIMED Consortium Data Sharing Agreement) before its respective MEMBER COMPONENT, DATA AFFILIATE COMPONENT, or NON-DATA AFFILIATE COMPONENT can join the PRIMED-SAG. Similarly, if a PRIMARY MEMBER or AFFILIATE withdraws from the PRIMED-SAG, any associated COMPONENTS will no longer be active (see also Term 7 of CDSA). Further, DATA AFFILIATE COMPONENT uploaders require approval from the Primary DATA AFFILIATE’s Investigator REPRESENTATIVE.
Human Genome Diversity Panel (HGDP)
Please see the PRIMED Steering Committee’s statement on the use of HGDP data within the PRIMED Consortium.
Back to topRelated Policies
- NHGRI Genomic Data Sharing (GDS) Policy: Data Standards
- PRIMED Consortium Code of Conduct
- PRIMED Core Membership Policy
- PRIMED Affiliate Membership Policy
- PRIMED Publications Policy
- Consortium Guidelines for AnVIL Data Access
- AnVIL Data Submission Guide
- dbGaP Security Procedures
Back to top
Change Log
- V3.1 - Consortium name updated
- V3.0 - Major policy update, approved on the August 21, 2024 Steering Committee call
- Added Definitions section and table of PRS Terms and Definitions
- Adding further detail on GSR sharing, and determination that PRS scoring files are GSR
- Added clarification on the “non-transferability” clause of the dbGaP Data User Code of Conduct
- Added new section on Access Removal, Withdrawal, and Termination
- Added detail and clarification to section on Data Use Guidelines pertaining to applicability of compliance with consent and limitations
- Linked to SC’s HGDP Statement
- V2.1 - Minor update to Figure 1 to depict the relationship of this DSP to the data sharing circles.
- V2.0 - Major policy update, approved on the November 15, 2023 Steering Committee call
- Clarifying management of the SAG, specifically relating to CDSA major version updates
- Adding determination from the NHGRI Data Sharing Governance Committee that PRS models are GSR
- V1.0 - Initial Policy, approved on the August 16, 2023 Steering Committee call.
